Direct sequencing of bacterial colonies by rolling circle amplification (RCA)
Direct Colony Sequencing (DCS) is an innovative technique that allows researchers to obtain high-quality genomic information directly from bacterial or fungal colonies. It bypasses the traditional process of isolating DNA from cultures, thereby saving time and resources while providing valuable insights into the genetic composition of individual microorganisms. DCS has revolutionized microbial genomics, enabling scientists to explore and understand the vast diversity of microorganisms present in various environments. This webpage explores the uses, value, and applications of Direct Colony Sequencing. Eurofins Genomics accepts the following sample types for direct colony sequencing:
- Bacterial Colonies
- Glycerol Stock / Bacterial Culture
- Bacterial Pellet
Rolling circle amplification process
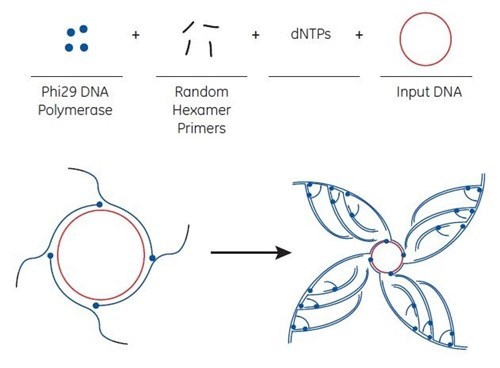
Rolling circle amplification (RCA) is a critical part of direct colony sequencing. It enables fast Sanger sequencing of plasmid clones without the need for plasmid preparation. The RCA method utilizes bacteriophage Phi29 DNA polymerase to exponentially amplify single- or double-stranded circular DNA templates. The isothermal amplification method produces microgram quantities of DNA from picogram amounts of starting material in a few hours.
An in vitro amplification of very small amounts of template DNA eliminates the need for overnight cell culture and conventional plasmid or M13 bacteriophage DNA purification. The proofreading activity of the Phi9 DNA polymerase ensures high fidelity DNA replication.
The Direct Colony Sequencing service has been validated primarily for plasmids in E. coli colonies. In principle, the method also works with large constructs (e.g. BACs) and other organisms. Contact us for pilot testing and optimization. As a starting point, we recommend performing cell lysis or crude DNA extraction and adding 1 to 5 µL of the lysate/extract to the buffer in the 96-well plates.
Value of Direct Colony Sequencing
Direct Colony Sequencing saves time. It sequences the plasmids/vectors directly from bacterial colonies without the need for plasmid preparation. You just need to pick the colonies and directly transfer them to one of the acceptable submission formats. The wells of the tube and/or plate contain buffer to preserve the colonies.
This fast and cost-effective solution can be used to quickly screen your DNA cloning experiments and select the candidate(s) for final confirmation via DNA preparation and sequencing.
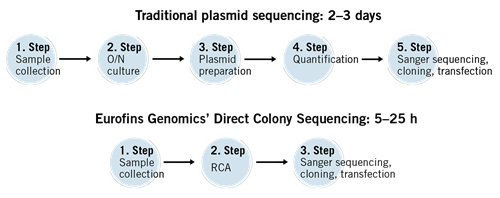
- Time and Cost Efficiency: DCS eliminates the need for DNA extraction and purification steps, reducing the time and cost associated with traditional methods. Researchers can directly sequence colonies, accelerating the genomic analysis process. This efficiency is particularly beneficial when dealing with large sample sizes or high-throughput applications.
- Single-Cell Genomics: DCS enables the genomic analysis of individual microbial cells. This is crucial for studying unculturable or slow-growing microorganisms that are difficult to isolate and cultivate. By obtaining genomic information at the single-cell level, researchers can uncover novel microbial species, discover new metabolic pathways, and gain insights into the genomic heterogeneity within microbial populations.
- Real-time Monitoring: DCS allows researchers to monitor the genetic changes occurring within microbial colonies in real time. By repeatedly sequencing colonies at different time points, researchers can track the evolution of microbial populations, study adaptive responses to changing environments, and identify genetic variations associated with specific phenotypic traits.
Applications of Direct Colony Sequencing
- Medical Research: DCS has numerous applications in medical research, such as identifying pathogenic microorganisms, studying antibiotic resistance mechanisms, and exploring the human microbiome. It aids in understanding the role of microbial communities in health and disease, facilitating the development of personalized medicine and targeted therapies.
- Environmental Studies: DCS is invaluable for investigating the diversity and functional potential of microorganisms in environmental samples. It helps in studying microbial interactions, nutrient cycling, and ecosystem dynamics. DCS can be applied to diverse environments, including soil, water, air, and extreme habitats, providing insights into the ecological roles of microorganisms.
Industrial Biotechnology: DCS plays a vital role in industrial biotechnology, enabling the discovery and optimization of microbial strains for the production
| Highlights |
Benefits at a glance |
| |
|
|
Fast
Receive your sequencing results one day earlier for your plasmid clones as DNA preparation is not needed. Results are available 15 hours after samples received.
Flexible
It is easy to submit direct colony sequencing online. Users can select the sample type from any of the standard sequencing submission pages online.
Cost-Effective
Reduce your expenses on plasmid DNA isolation and let us sequence your bacterial colonies directly.
|
- Available for E.coli colonies or any other gram negative bacteria
- Up to 4 reactions per sample, per plate are possible
- High quality read lengths of up to 1,100 bases
- Results are available 15 hours after samples received
- Free one time repetition in case of technical failure*
- Different sequencing result files can be selected per order
- A huge amount of standard primers are available free of charge
- Sequencing primers can be ordered or enclosed
- Easy re-order options from stored DNA samples
|
Uses of Direct Colony Sequencing
- Microbial Identification: DCS facilitates rapid and accurate identification of microbial species. By sequencing the DNA of colonies, researchers can compare the obtained sequences against databases to identify the microorganisms present in a sample. This can be particularly useful in clinical diagnostics, food safety testing, and environmental monitoring.
- Genomic Characterization: DCS provides a comprehensive view of the genetic makeup of individual microbial isolates. It enables researchers to analyze the presence of specific genes, genetic variations, and mobile genetic elements within a population. This information aids in understanding the functional potential, virulence factors, and antibiotic resistance profiles of microorganisms.
- Metagenomic Studies: Metagenomics involves analyzing the collective genomes of microbial communities present in a particular environment. DCS contributes to metagenomic studies by allowing researchers to selectively sequence individual colonies within a complex community. This approach enables a deeper understanding of the community structure, functional potential, and interactions among microorganisms.
- Phylogenetic Analysis: DCS facilitates the reconstruction of evolutionary relationships among microorganisms. By sequencing the 16S rRNA gene or other conserved genetic markers, researchers can generate phylogenetic trees to elucidate the evolutionary history of different species. This aids in taxonomic classification, identification of novel lineages, and understanding the diversity and evolution of microorganisms.
FAQ
- How is a technical failure defined?
- A technical failure is defined as a chemical or technical (IT, machines) defect in our lab. In order to ensure that the sequencing quality is not affected by a technical failure, an internal quality is run with each plate (for that purpose well H12 should be kept empty, otherwise a quality control is not possible). In addition to that, a sophisticated software is analyzing each plate considering parameters like quality values (QV) and reading lengths. If these parameters fall below a certain percentage across the plate an additional manual check is performed. If a technical failure is confirmed by this manual check, the plate is getting re-sequenced.
- What is the storage time?
- DNA and primer storage:
- Storage of template DNA for 4 weeks
- Storage of synthesized primers for 4 weeks free of charge
- Storage of synthesized primers for one year at additional cost
Data storage:
Secure online archive of all selected sequence data for 6 months.
Sample Prep
General Prep
- Shortly centrifuge the received plate to avoid the liquid on the foil. Carefully remove the sealing foil from the plate
- Grow your colonies on agar plates long enough to have a colony diameter of at least 1 mm*
- Use a toothpick to take as much of the bacterial colony as possible and inoculate into our plates. Swirl the toothpick for some seconds to transfer as many cells as possible into the wells
- In case of bacterial suspension use a pipette to add 5µl of the liquid culture in each well
- Well H12 should be kept empty for internal quality control
- Seal your plates using 8-cap strips to prevent material loss (8-cap strips are provided along with each PlateSeq Kit Colony)
- Plates can be sent / provided at ambient temperature to our sequencing lab
Sample Specific Prep
| Sample Type |
Required Prep |
Notes |
| Bacterial Colony |
RCA prep |
Numbered
Arrange colonies systematically in a grid formation on the agar plate and delineate those necessitating sequencing by encircling them. Assign numerical labels to each colony for identification purposes.
Unnumbered
Plates lacking numerical labeling will undergo processing as random picks. In the case of liquid culture, submit samples in a 96-well plate with strip caps.
Ensure the secure packaging of agar plates by wrapping them with parafilm and including a cushion to safeguard against potential damage during transportation. |
| Glycerol Stock / Bacterial culture |
RCA prep |
Tubes or 96-well plates are acceptable submission formats. Dry ice is required for shipping samples. Kindly label each with the type of media and any other useful information for the lab. |
| Bacterial Pellet |
Mini Prep |
Samples should be submitted as cell pellets from either BSL1 and BSL2 strains. We recommend resuspending in esuspended in Zymo 1X DNA/RNA Shield, and strongly discourage using alternative preservation media, such as RNAlater, given their suboptimal efficacy in DNA preservation and incomplete inactivation of host cells, which effects lab safety. We do not currently accept cell pellets from BSL3 strains.
Samples are best harvested from a fresh clonal culture while the bacteria is in an exponential growth phase or early stationary phase. Sending aged cultures in the death phase is strongly discouraged. |
Primers
Benefit from the greatest flexibility to handle your sequencing primers.
Thanks to our DNA synthesis lab, bioinformatic know how and internal IT solutions you can choose how to provide us with your sequencing primers:
Select from standard primers on stock
Select and manage your favorite standard primers from more than 80 different validated standard primers
Order specific sequencing primers
Specific primers can be ordered directly with your sequencing order. These primers are stored for 4 weeks and can be re-used during this time
Enclose your own sequencing primers
You can send us your sequencing primers in 1.5ml tubes along with your samples by using our Primer Barcodes*
Please consider optimal primer conditions, concentration and volume as described below.
Optimum primer conditions:
- Primers must not contain phosphorylation or fluorescent dyes
- The optimum primer length is between 16-25 bases
- The primer melting temperature (Tm) should be 50 - 62°C
- The GC content of the primer should be 35-60%
- Ideally one G or C should be located at the 3' primer end
- The number of 3' Gs or Cs should not exceed 2 Gs or Cs
- If possible, avoid >3 identical bases in a row in the sequence
Primer concentration and volume:
- Exactly 10 pmol/µl primer concentration is required per sequencing reaction
- Each primer must have a total volume of 15 µl (double distilled water or 5mM Tris-HCl); 5 µl of primer volume is required for every additional sequencing reaction
- Concentration of primers with wobble bases must be calculated according to the following formula: nX x ConcPrimer
n = number of bases within a wobble according to IUPC code, X = number of wobbles within the primer sequence. [e.g. 1 V (AGC) = 31 x 10 pmol/µl; 2 V (AGC) (AGC) = 32 x 10 pmol/µl]
*: Purchased Tube Labels can still be used to identify enclosed primers
Storage Times
DNA and primer storage:
- Storage of template DNA for 4 weeks
- Storage of synthesised primers for 4 weeks free of charge
- Storage of synthesised primers for one year at additional cost
Data storage:
Secure online archive of all selected sequence data for 6 months
Failures
Definition of technical failure
A technical failure is defined as a chemical or technical (IT, machines) defect in our lab. In order to ensure that the sequencing quality is not affected by a technical failure, an internal quality is run with each plate (for that purpose well H12 should be kept empty, otherwise a quality control is not possible). In addition to that, a sophisticated software is analysing each plate considering parameters like quality values (QV) and reading lengths. If these parameters fall below a certain percentage across the plate an additional manual check is performed. If a technical failure is confirmed by this manual check, the plate is getting re-sequenced.
Introduction
You have prepared your DNA construct, cloned it into a vector and transformed E. coli with it. Now it is time to pick and grow the colonies and conduct minipreps in order to extract plasmids to send for sequencing. Confirmation of a construct’s presence and identification of the construct in the vectors is absolutely essential before you continue to work with the plasmids. However, this process is quite laborious and time-consuming, especially when there are a large number of colonies to check.
Rolling circle amplification process
The basis for the Direct Colony Sequencing solution is the rolling circle amplification (RCA) technique that generates multiple copies of circular DNA. The starting material for amplification can be of various sources, such as from a small number of bacterial cells containing a plasmid, isolated plasmids and intact M13 phages to any circular DNA (including ligation reactions).
Bacterial colonies can be picked from agar plates and added directly to the 96-well plate. Alternatively, microlitre quantities of a saturated bacterial culture or a glycerol stock can also serve as starting material.
Process
RCA is performed at 30°C without thermal cycling. Depending on the source of starting material, amplification is completed in 6 to 18 hours. The amplification products are double-stranded concatemers of the circular template with high molecular weight. Concatemers are large DNA molecules that contain many copies of a specific DNA sequence.
For RCA, random hexamer primers anneal to the circular single-strand template DNA at multiple sites. The Phi29 DNA polymerase extends each of these primers. When Phi29 reaches a downstream extended primer, strand displacement synthesis occurs. Here, the original single-stranded DNA is replaced by the newly replicated strand. The displaced strand is then available to be primed by more hexamer primer. The process results in exponential amplification.
Note: when starting with M13 bacteriophage clones, the product is double-stranded DNA and can be sequenced with forward and reverse primers. DNA amplified by the method can be used directly without any purification.

The Direct Colony Sequencing service has been validated primarily for plasmids in E. coli colonies. In principle, the method also works with large constructs (e.g. BACs) and other organisms. Contact us for pilot testing and optimization. As a starting point, we recommend performing cell lysis or crude DNA extraction and adding 1 to 5 µL of the lysate/extract to the buffer in the 96-well plates.
References
Dean, F. et al., Genome Research 11, 1095–1099 (2001).
Lizardi, P. et al., Nat. Genet. 19, 225–232 (1998).
Estaban, J.A. et al., J. Biol. Chem. 268, 2719–2726 (1993).