High throughput microbial classification down to the species-level
INVIEW Microbiome High Specificity applies an amplicon-based targeted resequencing method based on PacBio’s SMRT sequencing technology.
INVIEW Microbiome High Specificity is a next-generation sequencing service that offers characterization of microbial communities down to the species level based on full-length 16S rRNA gene profiling. The product combines library preparation, sequencing with PacBio long read technology and reliable BioIT analysis for better separation of species in microbiomes that are difficult to resolve.
Specific applications of INVIEW Microbiome High Specificity include
- Identification of disease-causative agents and pathogens
- Species-level chacterisation of bacteria that are unculturable or biochemically unidentifiable
- Routine detection of medically important bacteria
- Reference identification for unusual strains
Highlights of INVIEW Microbiome High Specificity
- Excellent taxonomical resolution due to high throughput full-length 16S amplicon sequencing
- Classification of microbial communities down to species level
INVIEW Microbiome High Specificity is a sample-to-data package that offers
- Processing of up to 10 samples per run in parallel
- Amplification of whole 16S rDNA by PCR
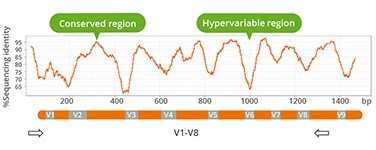
- Use of validated primers (V1-V8)
- Purification of PCR products
- Barcoding of PCR products, pooling and standard adapter ligation
- Library purification and QC
- Sequencing using PacBio technology
- Expert BioIT analysis for a comprehensive Data Analysis Report
- Available under ISO17025 accreditation
Accepted starting material for INVIEW Microbiome High Specificity
- >10 ng DNA with at least 10e3 bacterial/fungal genomes per sample and PCR amplification
- DNA isolation available as additional service for sand, soil, sludge, stool, urine culture, blood culture, milk, yoghurt (<3.5% fat), buccal swap, skin swap and tracheal fluid
Please note, that only S1-classified material is accepted for DNA isolation ordered online. More information about current classification of biological material can be found here (TRBA 466). Please contact us for further information on DNA isolation from other strating material or material classified as S2.
Specifications
Sequencing Technology
PacBio SMRT-Sequencing
Bioinformatics analysis
- Quality filtering of sequencing reads
- Removal of chimeras
- Ribosome Database Project-based OTU assignment (BLAST)
- Determination Shannon and Simpson diversity indices
- Combined OTU analysis
- Filtering and consolidation
Deliverables
- Raw data (fastq, bas.h5, metadata.xml)
- Sorted "Reads of Insert" (fastq)
- Non-chimeric and quality filtered sequences (fasta)
- Sequences w/ and w/o OTU assignment (fasta)
- Read statistics (tsv)
- OTU tables (tsv) and OTU distribution plots (png)
- Shannon diversity index tables (tsv)
- Simpson diversity index tables (tsv)
- Diversity index plots (png)
- Comprehensive Data Analysis Report (pdf)
Sequencing data delivered
- All raw sequencing data (.fastq)
- Tables listing all genera present in the analysed sample and the respective read counts
- Diversity indices
- Data delivery in common file types (.tsv, .txt), which can be handle with standard software for further analysis or visualisation
- Visualisation of relative species abundance
Delivery time
- 29 days for up to 10 samples
- Express delivery available: 21 days for up to 10 samples